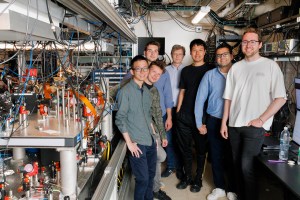
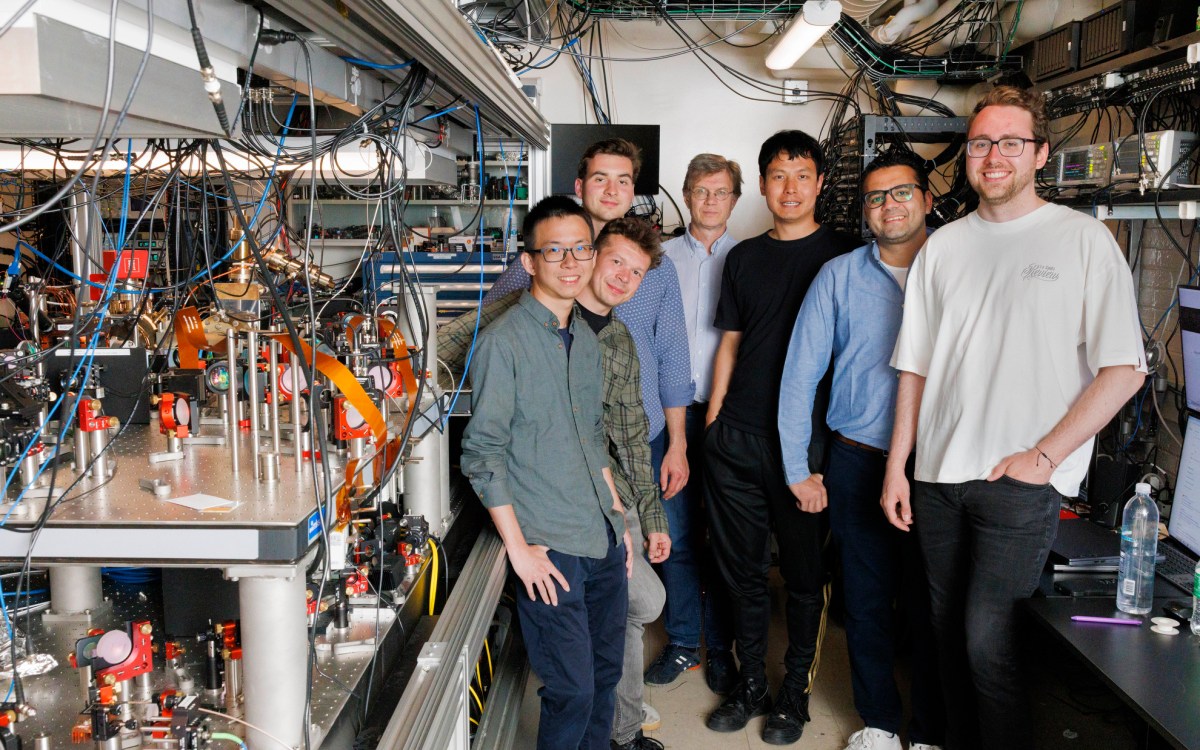
“Using twin prime editing is like using two word processors at the same time to simultaneously write different parts of the same paragraph,” explains Harvard Professor David Liu, the paper’s senior author.
Stephanie Mitchell/Harvard file photo
Twin gene-editing system gives twice the efficiency
New technique will allow programmable manipulation of large DNA segments
A team of researchers led by Harvard and Broad Institute scientists has developed twin prime editing, a new, CRISPR-based gene-editing strategy that enables manipulation of gene-sized chunks of DNA in human cells without cutting the DNA double helix.
Because it can make larger edits than previously possible, the new technique could make it possible to study and treat genetic diseases arising from loss of gene function or complex structural mutations, such as hemophilia or Hunter syndrome.
In the paper, published in the journal Nature Biotechnology, the researchers detail how they developed twin prime editing (PE), which uses a prime editor protein and two prime editing guide RNAs (pegRNAs) for the programmable replacement or excision of DNA sequences at endogenous human genomic sites without requiring double-stranded DNA cuts.
When combined with a site-specific recombinase, twin PE allowed integration of gene-sized DNA of more than 5,000 base pairs into the human genome at sites chosen by the researchers. Because large structural variants are found in many human disease-causing alleles, this technique may help treat genetic diseases. The study used twin PE plus recombinase to edit genes linked to Hunter syndrome, phenylketonuria (PKU), Duchenne muscular dystrophy, and hemophilia.
“A major remaining challenge in mammalian cell gene editing is our inability to make targeted gene-sized insertions at sites of our choosing,” said David Liu, the paper’s senior author, Thomas Dudley Cabot Professor of the Natural Sciences, and a core faculty member of the Broad Institute. “Such a capability could advance gene therapy by enabling genes to be restored in their native sequence locations, without increased risk of cancer from semi-random or uncontrolled integration at other locations in the genome.”
This work was performed by members of Liu’s lab, including former postdoctoral fellow Andrew Anzalone, former graduate student Chris Podracky, postdoctoral fellow Xin (Daniel) Gao, and current graduate student Andrew Nelson.
This new method is designed to overcome some of the limitations of existing gene-editing techniques, and build on the strengths of prime editing.
In 2016 the Liu group developed base editing, an efficient and precise genome-editing method that functions like a genetic pencil, chemically rewriting one DNA base to another without completely breaking the DNA backbone. The Liu group developed two classes of base editors that can correct four of the most common kinds of single-letter mutations, which collectively account for about 30 percent of known disease-associated human genetic errors. Three years later, the group developed prime editing, in which a reverse transcriptase directly copies edited DNA sequences into a specified target site from an extended guide RNA, again without requiring double-stranded DNA breaks. Like a genetic word processor, prime editing lets researchers search for one DNA segment and swap it for another.
Now, twin PE lets researchers search and replace larger DNA segments, again without breaking double-stranded DNA.
“Using twin prime editing is like using two word processors at the same time to simultaneously write different parts of the same paragraph,” said Liu, who is also a Howard Hughes Medical Institute Investigator. “In the process, you can create that paragraph more efficiently.”
The researchers developed twin PE by using simultaneous prime edits to create both strands of the edited DNA. By itself, twin PE enables efficient deletions, insertions, and substitutions of hundreds of base pairs, achieving larger edits than simple prime editing. Seeking the ability to make even larger edits of up to thousands of base pairs — a size large enough to include many whole genes — the team installed Bxb1 recombinase “landing sites” at targets site in human cells. The recombinase then mediated the insertion of large DNA cargo sequences into the landing sites.
The team tested twin PE plus Bxb1 recombinase by inserting large DNA into a variety of target sites in the human genome, including at “safe harbor loci” thought to be ideal for gene therapy because inserting genes there has been found to not induce cancer or other apparent toxicities.
The researchers also applied twin PE-mediated deletions to target DMD. Pathogenic DMD alleles, which cause Duchenne muscular dystrophy, commonly contain large deletions in exonic regions that result in frame-shifted transcripts. These experiments show that twin PE can generate large deletions at therapeutically relevant loci in humans with far fewer potentially harmful byproducts than paired Cas9 nuclease strategies.
The team also used twin PE and a recombinase to install a pathogenic 40,000 bp inversion that causes Hunter syndrome. Their success suggests that the combination might eventually work as a therapeutic strategy for correcting other large or complex pathogenic gene variants.
Going forward, Liu is optimistic that other recombinases can work effectively with twin PE.
“We hope to apply twin PE to some basic science and some therapeutic questions, while continuing to develop this strategy to increase the efficiency of large gene size integrations,” Liu said.
Podracky, who joined the lab in 2015, said he hopes that the twin PE plus recombinase technique will both be studied and be an inspiration for scientists across the country.
“We could see other academic labs take the technology and put it put it to good use,” Podracky said. “If this enables [scientists] to realize their gene-editing ideas more readily, that would be a success.”
This study was supported by the Merkin Institute of Transformative Technologies in Healthcare, the National Institutes of Health, and the Howard Hughes Medical Institute.