The work of Harvard researchers was recognized in the “2018 Breakthrough of the Year” edition of Science magazine.
Photo by Amanda Swinhart
Breakthrough science recognized
Harvard teams’ studies featured in Science ‘Breakthrough of the Year’
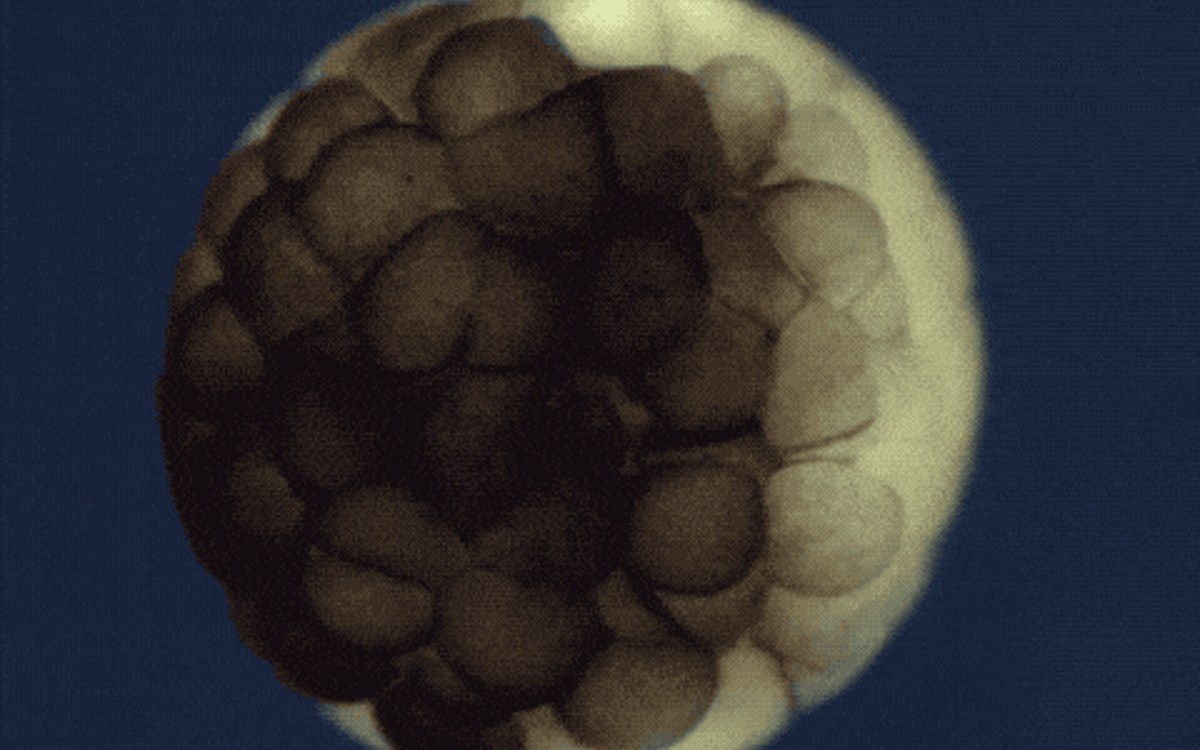
Two series of studies by Harvard research teams, who used a combination of technologies to track the development of zebrafish and frog embryos, cell by cell and through time, have been featured in the “2018 Breakthrough of the Year” edition of Science magazine.
One set of studies was led by Alexander Schier, the Leo Erikson Life Sciences Professor of Molecular and Cellular Biology at Harvard University. The other series was conducted by Harvard Medical School (HMS) researchers, led by Allon Klein, assistant professor of systems biology, Sean Megason, associate professor of systems biology, and Marc Kirschner, the John Franklin Enders University Professor of Systems Biology.
Their work was described in three papers published in Science in April and a paper in Nature Biotechnology in March.

The teams were able to reconstruct the developmental trajectories and lineages that generate neurons, muscle, blood, and other cell types in unprecedented detail, using a combination of technologies including single-cell RNA sequencing and CRISPR-Cas9 genome editing, and computational approaches.
Science editor in chief Jeremy Berg, in announcing the recognition, commented that the work addresses “one of the most fundamental and fascinating processes in biology — the seemingly miraculous transformation of single cells into complex organisms — providing rich information about cell-type inventories and laying the foundation for many future studies.”
“The large and rich data sets that have been generated, and the techniques that will produce more, constitute exciting breakthroughs in developmental biology,” Berg wrote.
The Harvard research teams traced the fates of individual cells over the first days of an embryo’s life, and revealed a comprehensive landscape of which genes were switched on or off, and when, as embryonic cells transitioned into new cell states and types.
Together, the findings represent a catalog of genetic “recipes” for generating different cell types in two vertebrate species and provide an unprecedented resource for the study of developmental biology and disease.
More like this
“With single-cell sequencing, we can, in a day’s work, recapitulate decades of painstaking research on the decisions cells make at the earliest stages of life,” said Klein. “With the approaches that we’ve developed, we’re charting what we think the future of developmental biology will be as it transforms into a quantitative, big-data-driven science.”
In addition to shedding new light on the early stages of life, the work could open the door to a new understanding of a host of diseases, said Schier.
“We foresee that any complex biological process in which cells change gene expression over time can be reconstructed using this approach,” he said. “Not just the development of embryos, but also the development of cancer or brain degeneration.”